 Venter
DNA Project
Venter
DNA Project
To understand many of the concepts associated with genetic genealogy, one must have an understanding of the basics of Genetics. It is the purpose of this discussion to help the reader develop an understanding of the fundamental genetic concepts and how they can be applied to genealogical research. Here we present the principles and define the terms you will come across in any discussion of genetic genealogy. The intent is to present the material in simple terms and not burden the reader with a detailed scientific dissertation. Many of the terms used may be new to the reader, but they are all defined and used in context, which will aid in understanding. All important terms are written in italics the first time they are used in the text.
The human body is made up of a complex arrangement of assorted parts, such as lungs, heart, stomach, brain, blood vessels and the like. But, each of those individual parts is made of living cells. And each of these cells are identical to each other in that within the nucleus of each is a copy of the complete blueprint for that particular human body which the cell inhabits. That blueprint is called the genome. What exactly is a genome? The genome is the total set of all the instructions which the body needs to grow and maintain itself. The genome supplies the cell with the information the cell needs in order to know what its assigned function is and how to perform that function. Different cells perform different functions, but they all get their instructions from the genome.
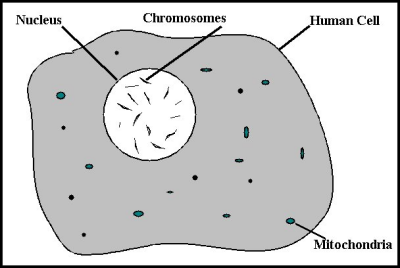
Figure 1: The Human Cell
The set of instructions which the genome contains is distributed across 23 pairs of chromosomes. One chromosome of each pair is received from the father and one from the mother. Each pair of chromosomes contains a specific sub-set of the instructions for the cell, with different sub-sets of instructions contained within each of the other chromosomes. All of the chromosomes taken together provide a complete set of instructions or, in other words, a complete genome. Individually, each instruction is know as a gene.
Of the 23 pairs of chromosomes, 22 have equivalent sets of genes on each of the chromosomes in the pair, one set from each parent. These first 22 pairs are known as autosomes. The 23rd pair is not really a pair at all, but two different chromosomes, one given the label X and the other labeled Y. The X- and Y-chromosomes are called the sex chromosomes and determine the gender of an individual. A female will have two X-chromosomes, while a male will have an X- and a Y-chromosome. Since females have two X-chromosomes a mother can only pass an X-chromosome to her child. Whereas, since males have both an X- and a Y-chromosome, a father can pass either his X- or his Y-chromosome to his child, resulting in an infant girl or boy, respectively. In this way it is the father, or more specifically, the father's sperm, that determines the gender of each child.
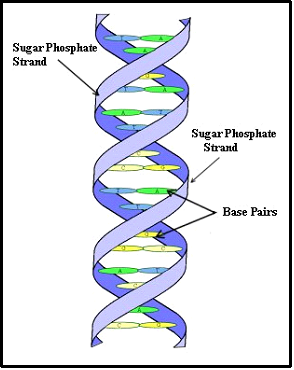
Figure 2: The DNA Molecule
A chromosome is a large assembly of molecules, which, together, form a molecule of DNA (deoxyribonucleic acid). The DNA molecule is very long, and is made up of smaller molecules stuck together in a specific way. It is the specific arrangement of these smaller molecules that encodes the instructions to the cell. These smaller molecules come in three types; phosphate, sugar, and base, and they bind together to form the famous 'Double helix' of the DNA molecule. One can visualize the DNA molecule as two long strands of alternating sugar and phosphate molecules with the base molecules attached to each strand and holding the strands together, much like the rungs of a ladder. See Figure 2. To say it another way, a base molecule attaches to a sugar molecule on one strand and a second base attaches to a sugar on the other strand and then the two bases attach to each other, forming a rung of the ladder. Each base molecule plus its phosphate/sugar group to which it is connected is called a nucleotide. The two bases, which attach to each other are referred to as a base pair.
A lot can be learned from understanding the exact sequence of all of the nucleotides in the complete genome, so the Human Genome Project was organized to decode it in its entirety. The Project, completed in 2003, was an intense international government and commercial effort, whose goal was to determine the exact sequence and identity of the more than 3 billion base pairs which make up the human genome and to identify the approximately 20,000-25,000 genes it contains. It was found that the genes make up less then 5% of the DNA in the genome. This percentage is referred to as the coding portion of DNA and is the portion that contains the coded instructions for the cell. The rest of the DNA, the non-coding portion, has no known purpose and is often referred to as junk DNA.
There are four base molecules involved in the making of DNA. They are adenine, designated by the letter A, thymine (T), guanine (G), and cytosine (C). It is not important to remember the names of each of these base molecules and, for our discussion here, they will just be referred to by their letter designator: A, T, G, or C.
Now, in building a DNA molecule, an A will only bind with a T, and G will only bind with C. So, if you know there is an A attached to the strand of phosphate/sugar on the left, you know that it will only bind with a T on the strand on the right. Therefore, knowing the order of bases, as you run along the left strand, allows you to determine immediately the order of bases on the right, because those on the right must be the complements of those on the left. This fact is what allows the DNA molecule to split and produce a copy of itself.
Let's take minute to review what we know so far? We know that all human cells contain a nucleus and each nucleus contains a complete genome. The genome is made-up of 23 pairs of chromosomes. Each chromosome is a long DNA molecule which contains a sub-set of the instructions needed by the cell to perform its function. The DNA molecule is comprised of phosphate and sugar molecules strung together to form two long strands, which are connected by pairs of base molecules. These base molecules are either a combination of A-T or C-G. The complete assembly resembles a ladder. That ladder is then twisted to form a double helix, as shown in Figure 2.
As noted above, DNA contains the instructions, known as genes, needed for the cell to function. The code for each gene is a specific sequence of bases (the nucleotides). Just as the letters of the alphabet are used to encode words, the four nucleotides are used to encode genes. The sequence of these nucleotides, in various combinations, allows for an almost unlimited number of possibilities. For example, these sequences could be such combinations as CTA, TTC, GATCTA, GTCA, and so on. A specific sequence of nucleotides will result in a specific instruction to the cell. Just as when you change the letters in a word (e.g., change 'make' to 'made' or 'take') you change the meaning of the word, so to, when you change the nucleotides in a gene (e.g., change CCA to CAA) you change the meaning of that gene. It is for this reason that the coping of DNA must be extremely accurate.
At the time of replication the DNA molecule is unzipped down the middle, with the break occurring between the two nucleotides of each base pair, splitting the pair as one phosphate/sugar strand peels away from the other. Then, two new strands are assembled, each one connecting to one of the old strands. Following the rule that As only bind to Ts and Gs only bind to Cs, each original strand acts as a template for the new copy. This results in two DNA molecules where there was originally only one. Replication is a very accurate process, which creates an exact duplicate time after time. However, on very rare occasions an error can and does occur. These errors are called mutations.
Mutations can occur at any time, anywhere on any chromosome, in any cell. But the mutations that are relevant to genealogy are only those that occur during the production of an egg or sperm cell and are called germ line mutations. Germ line mutations are the only mutations that can be passed on to the next generation. Mutations in all other cells are limited to the individual in which they occur. If a mutation occurs in the coding portion, a cell will not get the correct instruction and something unintended will occur. This type of mutation results in differences between individuals, such as hair or eye color, but can also cause genetic defects and inherited diseases. As a general rule, a mutation in the coding region is a disadvantage, and in most cases the defective gene does not survive in future generations. However, mutations occurring in junk DNA, those vast stretches of non-coding material, have no affect on survival whatsoever and, therefore, will be passed on from generation to generation, indefinitely.
Mutations that occur in the junk DNA will accumulate over time. They first occur in a single individual, known as the founding person for that mutation, and then will be passed on to succeeding generations. For those people with a specific mutation, that mutation is a signature of their ancestry and is unique to their lineage. All people with a given mutation must have descended from the founding individual.
The mutations that occur in the non-coding DNA can take several forms, but there are only two types that are important to genetic genealogy. The first type occurs when, in the process of replication, the original nucleotide is replaced by one of the other nucleotides. Let's say there was the sequence ATGC. Now, suppose an error was introduced and, for example, the G was replace by a T, resulting in the sequence ATTC. This is called a Single Nucleotide Polymorphism, or SNP for short. Single Nucleotide, because only one nucleotide location was affected, and Polymorphism, because this location in the DNA molecule can now exhibit either one or the other of these nucleotides in any given individual. The set of nucleotides that can appear at a given location is called the alleles for that location. In this case the alleles for this SNP are G and T, either of which can be found at that location. In addition to resulting from the replacement of one nucleotide by another, SNPs may also be the result of a deletion of an existing nucleotide or the addition of an extra one. A single nucleotide polymorphism is a type of mutation that is so rare, that the ones that are known to have occurred at a given location, undoubtedly have only occurred once in the history of the human species.
The second type of mutation that can occur is quite different from a SNP. In the long strands of DNA, short segments of nucleotides occur that are repeated over and over, five, ten, twenty times, or even more. However, the sequence that is repeated is usually only 1 to 6 nucleotides long. Let's say that the sequence is AGT, for example. If this sequence was repeated four times it would look like AGTAGTAGTAGT. Now there is something about having several repeats like this that causes the DNA replication mechanism to get confused. You could say that it loses its place during the act of coping, and occasionally it ends up making either too many repeats or not enough. Interestingly, the actual sequence of AGT is accurately reproduced, but the number of repeats is not. So, where we originally had four repeats of the sequence, we end up with 3 or 5, or maybe even some other amount. These stretches of repeated sequences are known as Short Tandem Repeats (STRs) and there are perhaps thousands of them scattered throughout the genome. Getting the number of repeats wrong during replication is a type of mutation that is much more common than a Single Nucleotide Polymorphism. In fact, STR mutations can occur as often as 2 times in a thousand. And unlike SNPs, where there is either one of two possible values, STRs are very polymorphic, and come in a variety of alternative versions (alleles), based on the number of repeats.
Twenty-two pairs of chromosomes within the cell nucleus are autosomes. Autosomes refer to chromosome pairs in which one chromosome from each pair was received from the father and one from the mother. An important aspect of autosomes is that in each parent, just prior to the creation of a germ line cell (e.g., an egg cell, in the case of the mother, and a sperm cell in the case of the father), the two chromosomes of each pair split apart and exchange some, but not all, of their genes. So, the chromosome a person gets from his or her mother is actually a mixture of the genes that the mother got from her two parents. The same is true for the other half of the chromosome pair which was received from the father. This process is called recombination and results in genes being constantly mixed through the generations.
But the Y-chromosome is not an autosome. It doesn't come in pairs, and, therefore, it doesn't have anything to recombine with. The Y-chromosome is also special in that it is the chromosome that determines gender. A man has an X- and a Y-chromosome and if he passes the Y on to his child, that child will be male. So a Y-chromosome is not only uniquely male, but a father passes it, unchanged, on to his son, who likewise passes it unchanged on to his son, ad infinitum. If it wasn't for an occasional mutation, every man would have exactly the same Y-chromosome. Y-chromosome mutations are important to genealogy because any mutations that occurs will follow paternal family lines just like the surname does. In fact, a specific set of mutations on the Y-chromosome can be considered the family genetic signature.
This is an important feature of genetic genealogy and because of it the following discussion will focus on the male line and the Y-chromosome. Analysis of the Y-chromosome is done by looking at specific locations on the chromosome which exhibit one of two kinds of mutations; Single Nucleotide Polymorphism and Short Tandem Repeats.
If you take any two people from different locations around the world, their DNA will be ~99.9% the same. This leaves about 3 million differences . Most of the differences are the result of Single Nucleotide Polymorphisms and most of the SNPs are located in the junk DNA. SNPs are used primarily in Anthropology to aid in the study of the Deep Ancestry relationships of human populations. They were chosen for this purpose because they are so rare and their frequency of occurrence differs throughout the world. This allows them to be used to define lineages known as haplogroups. A haplogroup is a cluster of people who share the same mutation, which they all have inherited from a single, common ancestor. The mutation that defines a haplogroup is given a kind of cryptic label such as M89, M343, or P25. These labels are not meaningful and one should not try to derive any sense from them, just know that each haplogroup is defined by a mutation with such a label.
An SNP originates in a single founding individual, and is then passed on to succeeding generations. Over time some of the founder's descendants may migrate away from the place where the mutation first occurred, but the location where the mutation originated is usually the place where the highest concentration of men with that mutation will tend to be found. So, taking into account the age of various mutations and the frequencies at which they occur in the locations where they are found today, will allow one to map the migrations of human populations across time and distance.
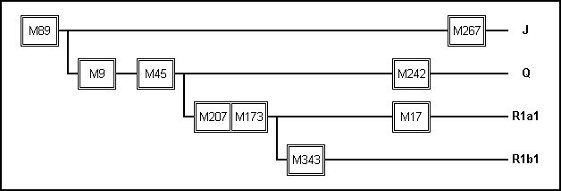
Figure 3: A Portion of the Phylogenic Tree for Y-chromosomes
Sometime around 45 thousand years ago a male child was born, probably in northeast Africa , and a certain SNP mutation had occurred on his Y-chromosome. Not to many years after that, a band of some of his descendants migrated out of Africa into the Middle East. And the descendants of this small group of wanderers eventually went on to populate the entire world, resulting in 90 - 95% of the worlds population today. That mutation which they carried has been labeled M89. Then, sometime around 40 thousand years ago, there was another birth, probably in Iran or central Asia , with another SNP, which has been labeled M9. At that time most of the men of the world had the M89 mutation and now the descendants of this second child will have both the M89 and the M9 mutations. In the following millennia other mutations occurred in other male children and the total population became diversified with various lineages having specific sets of the assorted SNPs. These lineages can be arranged into a kind of family tree of the human race known as a Phylogenetic Tree, a small portion of which is shown in Figure 3. Each branch of the tree represents a haplogroup (also called a clade) and is differentiated from its ancestor by a single SNP. From Figure 3, it is easy to see the relative age of each SNP and the order of its occurrence. Figure 3 shows the derivation of four haplogroups; J, Q, R1a1 and R1b1 (noted on the right side of the chart). However, each mutation on the tree actually represents its own haplogroup. For example, the mutation M89 founded the F haplogroup and M207 founded the R haplogroup. The four haplogroups noted on the right side of the diagram descended from these earlier haplogroups. The individual names of the haplogroups where assigned in alphabetical order and so have no meaning outside of their use as a label.
The R1b haplogroup (resulting from SNP M343) is the oldest in Europe and is the most common in western Europe. It was part of a mass migration into Europe around 20 to 30 thousands years ago and these people are the direct descendants of the Cro-Magnons. The R1a1 haplogroup (SNP M17) originated much later, around 10 thousand years ago, probably in present-day Ukraine or southern Russia. It came west from the steppes of southern Russia into central Europe and today can be found from western Poland east through Latvia and Belarus, into western Russia. Spenser Wells says that "Today a large concentration - around 40 percent - of the men living from the Czech Republic across the steppes of Siberia, and south throughout central Asia are part of this group".
A Haplogroup, as defined by an SNP, is like having a group of people with the same surname, but without knowing any of the first names. As a result, there is no way to distinguish people within the group. What is needed for genealogy is something the defines differences within the haplogroup, and can be used to track discrete family lineages. This requires a genetic mutation the changes relatively rapidly and Short Tandem Repeats fulfill this need very nicely.
As we know, STRs are short sequences of nucleotides that are repeated a number of times. The specific location at which an STR is found on the Y-chromosome is called a marker. In genetics, the term marker is used in the sense of a landmark, benchmark, or milepost. In the literature, one can also find these markers referred to as Microsatellites. When used for genealogical purposes, specific markers are chosen where the number of repeats are found to have considerable variation among individuals, making it an easy way to determine relatedness. STR markers are identified by a DNA Y-chromosome Segment (DYS) number, such as DYS #392 or DYS #385b, to name just two. When a Y-chromosome is analyzed, it is the number of repeats that is counted and reported for each marker site. For instance, marker DYS #392 generally has allele values that fall in the range of from 7 to 16. That is, the number of times the nucleotide sequence is repeated at site DYS #392 will usually be anywhere from 7 to 16.
When the Y-chromosome is tested commercially for genealogical purposes, a test for at least 12, and as many as 67, markers may be done. Which specific markers and the total number analyzed will vary from lab to lab. The results are usually report as a string of the marker allele values in a set order. If the test found the DYS #392 had 12 repeats, DYS #385a had 11 repeats, DYS #385b had 14 repeats, and DYS #390 had 24 repeats, the results would be reported as the series "12, 11, 14, 24". These results, taken as a group, are called a haplotype. A haplotype is simply an individual's complete set of results for whatever markers were tested. Unless two men are very closely related, their haplotypes will most likely differ by some amount. The degree of difference will depend on the number of markers tested and how closely the two men are related. The more markers, the more chance of differences; the more closely related, the less chance of differences.
A few examples will give you an idea of how these two factors interrelate. The haplotype of two brothers, cousins, or a father and son should have complete agreement, no matter how many markers are tested. Although, keep in mind that mutations do occur and have to occur somewhere, but, in general very closely related men will have identical haplotypes. Now, two men, with a common ancestor ten generations back, may differ by one, or possibly two markers in 24, and maybe even one in 12. But, showing more than a 3-marker difference in 24 would probably indicate there is not a common ancestor within the time period of most genealogical research (e.g., say 500 - 600 years). On the other hand, a five marker difference in a 67-marker comparison, would not be uncommon for men with a common ancestor 10 generations back ( say 250 - 300 years).
The field of Genetics is important to genealogy because it provides a foolproof way of tracing ancestral lines of individuals and groups of people where a paper trail doesn't exist. Through Single Nucleotide Polymorphisms, a man can determine his Deep Ancestry, tracing his ancestors across the globe, back to the first band of wanders to leave Africa almost 50 thousand years ago. And Short Tandem Repeats will allow him to identify distant members of his own family, although they may be separated by vast distances as well as a number of generations back to a shared ancestor.
© R. J. Venter, January 2009
Return to Top of page.
Return to More About DNA
Visit the Venter Surname Research Center Home Page.
Visit Robert Venter's Genealogy Home Page.
Prepared by Robert Venter
Venter Surname Project Leader
 Send e-mail to: venter.genealogy@comcast.net
Send e-mail to: venter.genealogy@comcast.net
Unsere deutsche Freunde dürfen uns auf Deutsch schreiben.